The ABiMS platform aims to develop and provide infrastructures for analysis purposes. Those e-Infrastructures are composed of different types of components:
The Hardware is mainly composed around a High Performance Computer constituted of 1800 CPU. Jobs are submitted through a scheduler (SGE). Other layers can be added like virtualization servers to host applications on dedicated and isolated virtual machine (VM).
Some public Databanks like genomics resources (GenBank, Uniprot, …) are provided (/db/) to different applications (CLI software, Galaxy workflows) in several indexing formats (Blast, Bowtie, BWA, …). Most of them are automatically downloaded and formatted using BioMAJ.
The platform, since 2012, is strongly involved in the Virtual Research Environment (VRE) Galaxy. ABiMS takes part in different national initiatives (with the working group IFB-Galaxy) and international initiatives (with the Elixir Galaxy Group). The platform takes part also in the organisation of workshops and conferences (GCC2017) and in the Galaxy developments with the project Workflow4Metabolomics, the Galaxy Training Materials, with some hacks in IUC Tools, and some minor commits in Planemo and Galaxy itself.
Furthermore, we maintain several open access Galaxy instances (see: Resources / Galaxy)
We also propose, through collaborations, some genome browsers (JBrowse) and collaborative genomic annotation editors (Apollo) from the GMOD project.
EOSC Life project (2019-2023) project
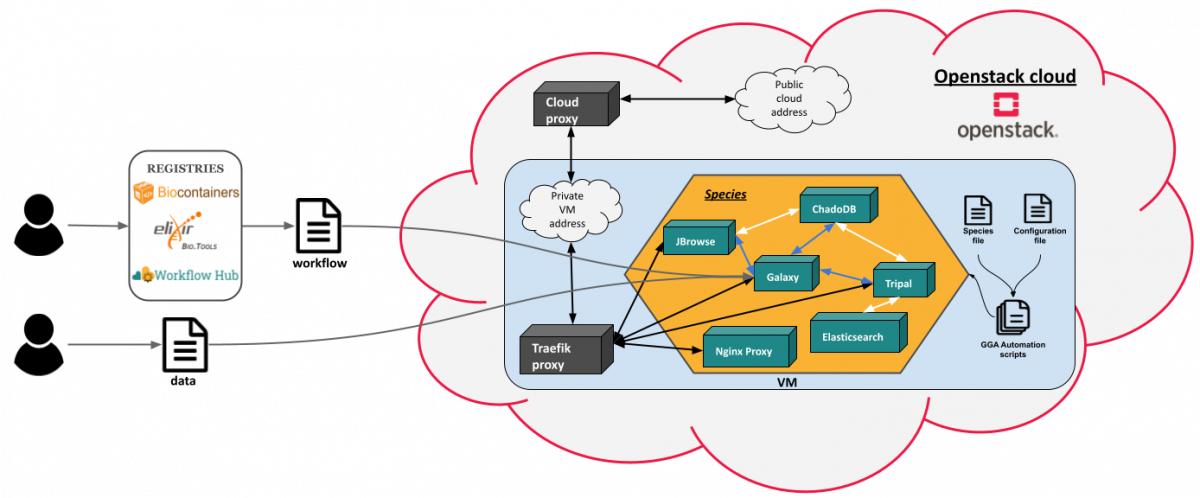
The EOSC-Life project aims to create an open collaborative digital space for life science in the European Open Science Cloud (EOSC). The ABiMS bioinformatics platform, member of the European Marine Biological Resource Centre (EMBRC) research infrastructure, is involved in the work package 2 (WP2) dedicated to make computational tools, workflows and registries findable, accessible, interoperable and reusable (FAIR). As part of the EOSC-Life project, we have added a new use case for genome annotation using genome synteny relationship into the GGA resources and implemened the GGA environment in the EOSC:
- We contributed to the demonstrator 4 use case developed by the CCMAR that was addressing the transfer of genome annotations between closely related marine species – as a test case, pelagic fishes - using genome synteny relationships. In this context, we have integrated necessary tools into Galaxy and the Toolshed (LAST tools for genome comparison and alignment, gffalign to for gene extraction and annotations transfer using genome synteny), and we have developped a Galaxy Docker image containing the workflow tools, available on quay.io.
- We have also developped Ansible recipes to allow the deployment of a Virtual Machine in a an Openstack cloud via the Terraform software, the installation of the GGA environment and its dependencies, as well as loading data into the Galaxy library. The Galaxy Genome Annotation (GGA) project consists of several projects and tool suites that are working closely together to deliver a comprehensive, scalable and easy to use Genome Annotation experience.